is a user-friendly and Galaxy-supported pipeline allowing to analyse large sets of DNA amplicons sequences accurately and rapidly.
is a user-friendly and Galaxy-supported pipeline allowing to analyse large sets of DNA amplicons sequences accurately and rapidly.
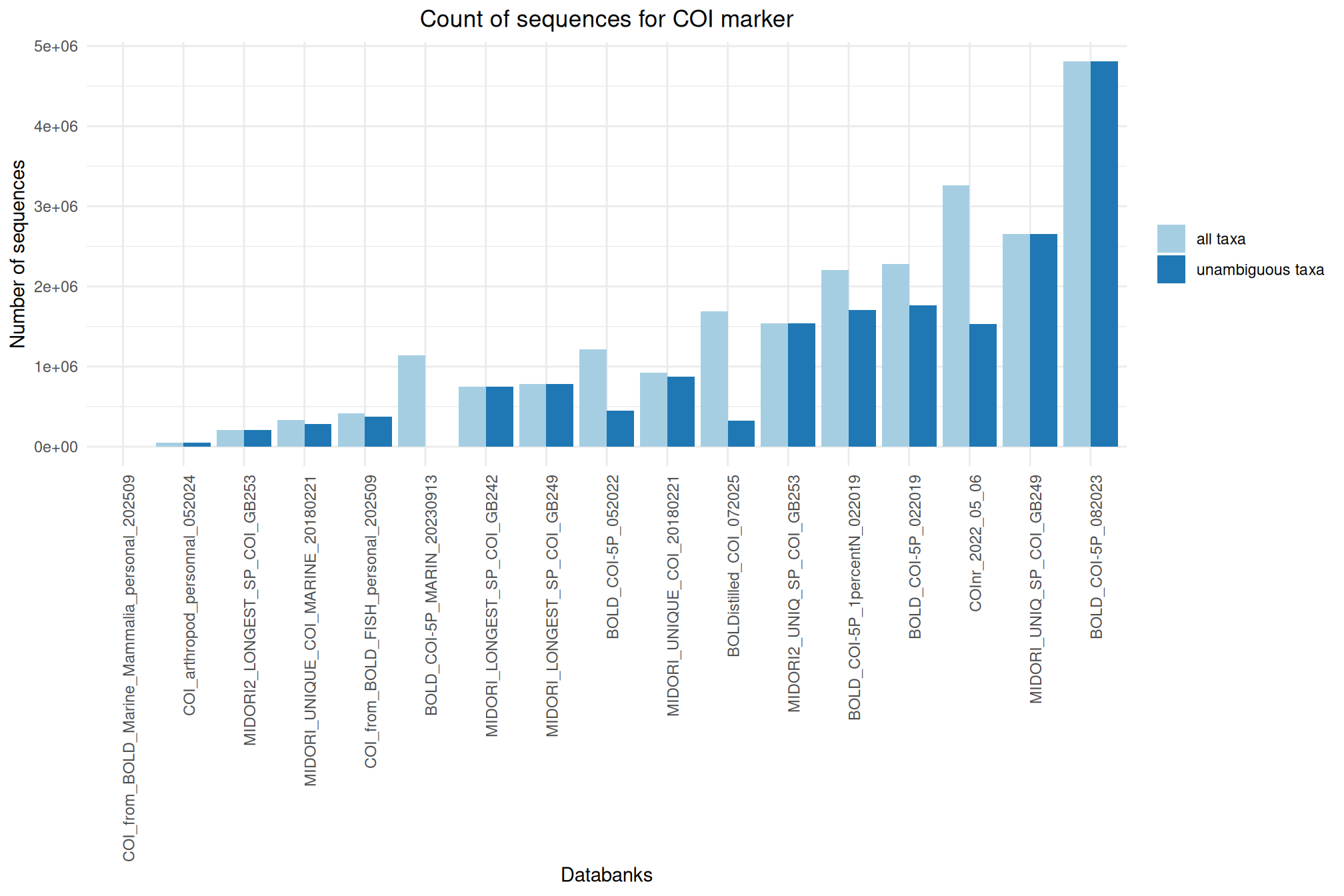
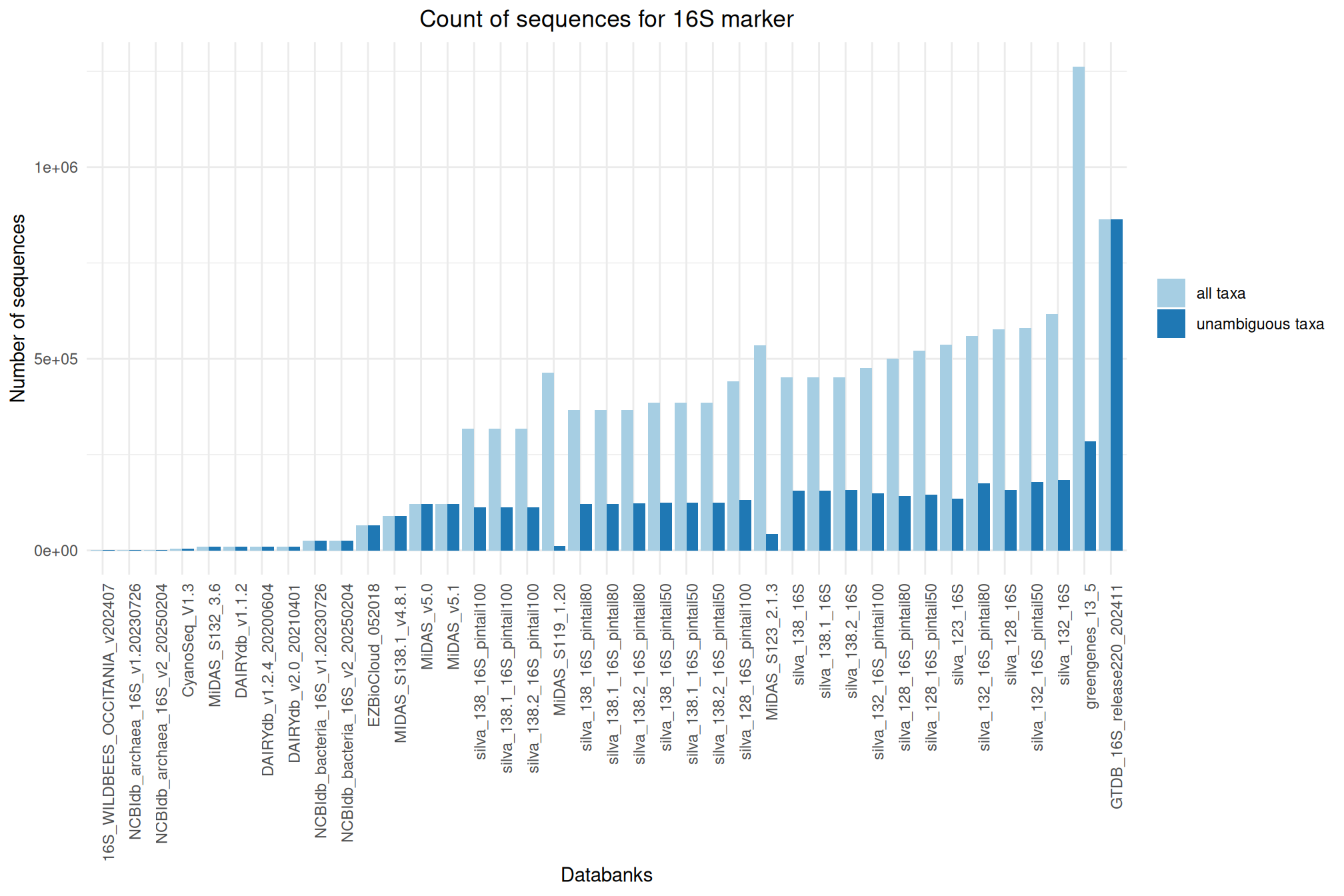
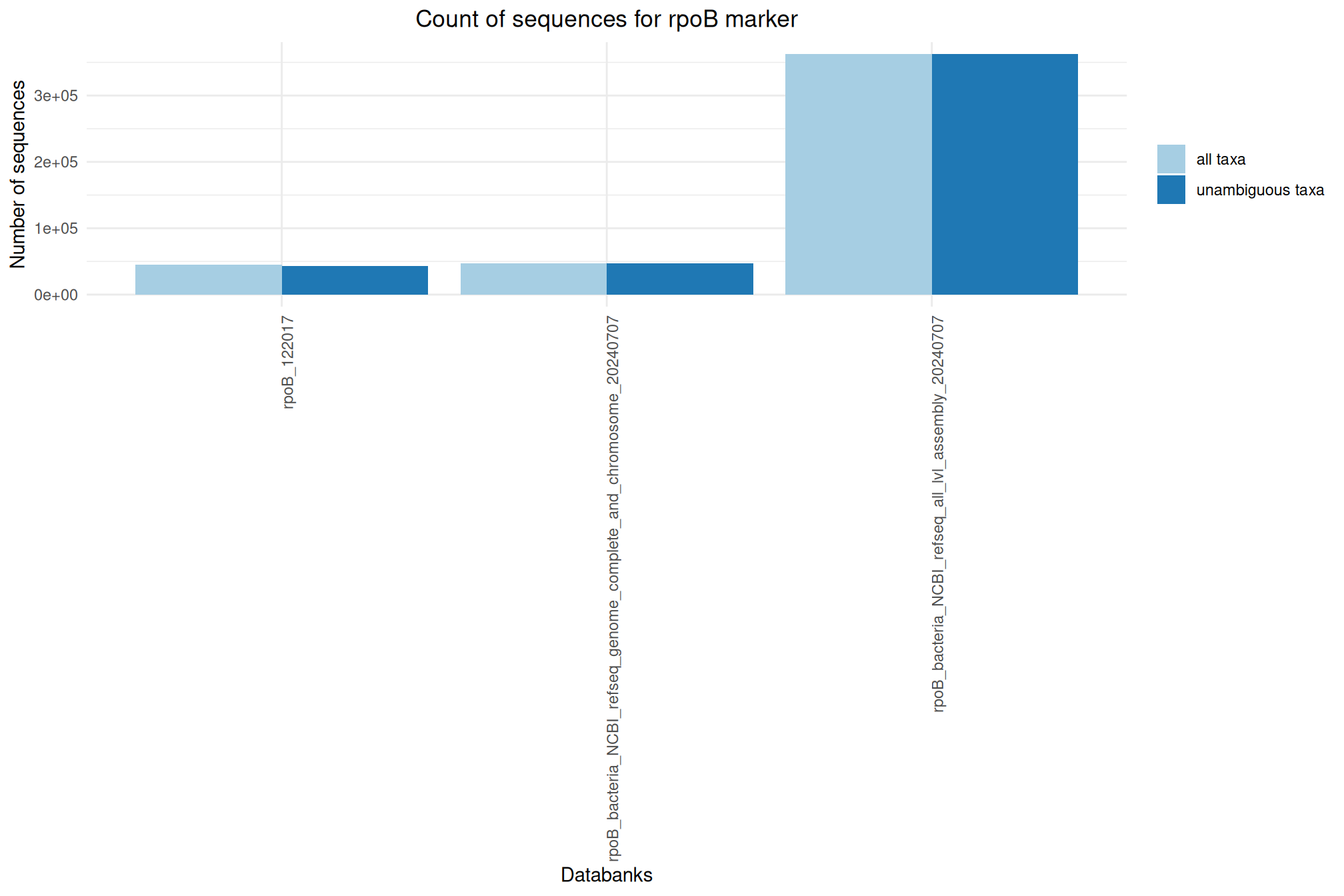
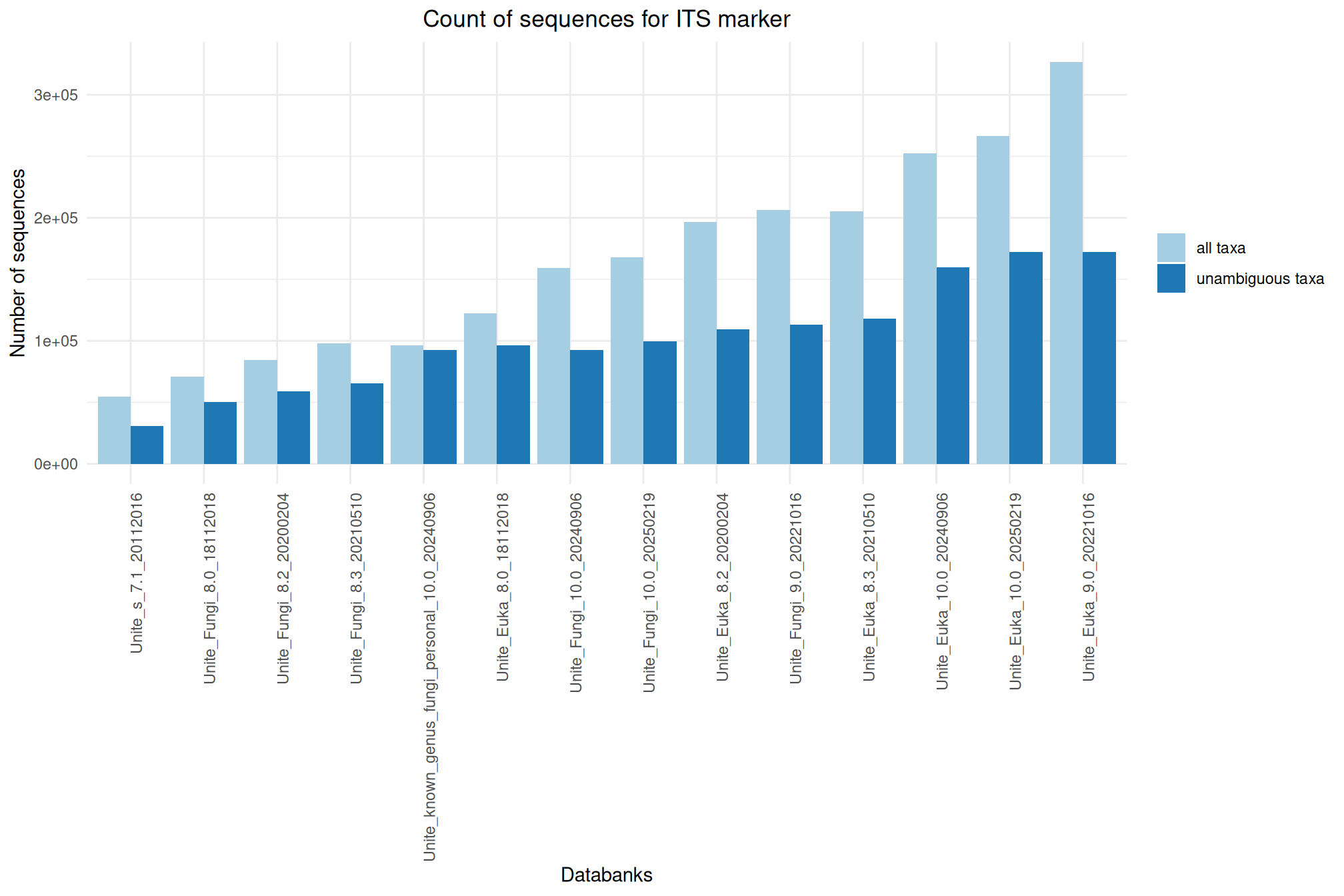
It offers numerous databanks for the affiliation step. We’ve put together some information and metrics to help you choose the most suitable databank for your analyses.
Dashboards
| Name | Date | Marker | Sequence number |
|---|---|---|---|
| BOLDistilled_COI_072025 | Sep 1, 2025 | COI | 1690200 |
| COI_from_BOLD_FISH_personal_202509 | Sep 1, 2025 | COI | 416693 |
| COI_from_BOLD_Marine_Mammalia_personal_202509 | Sep 1, 2025 | COI | 3397 |
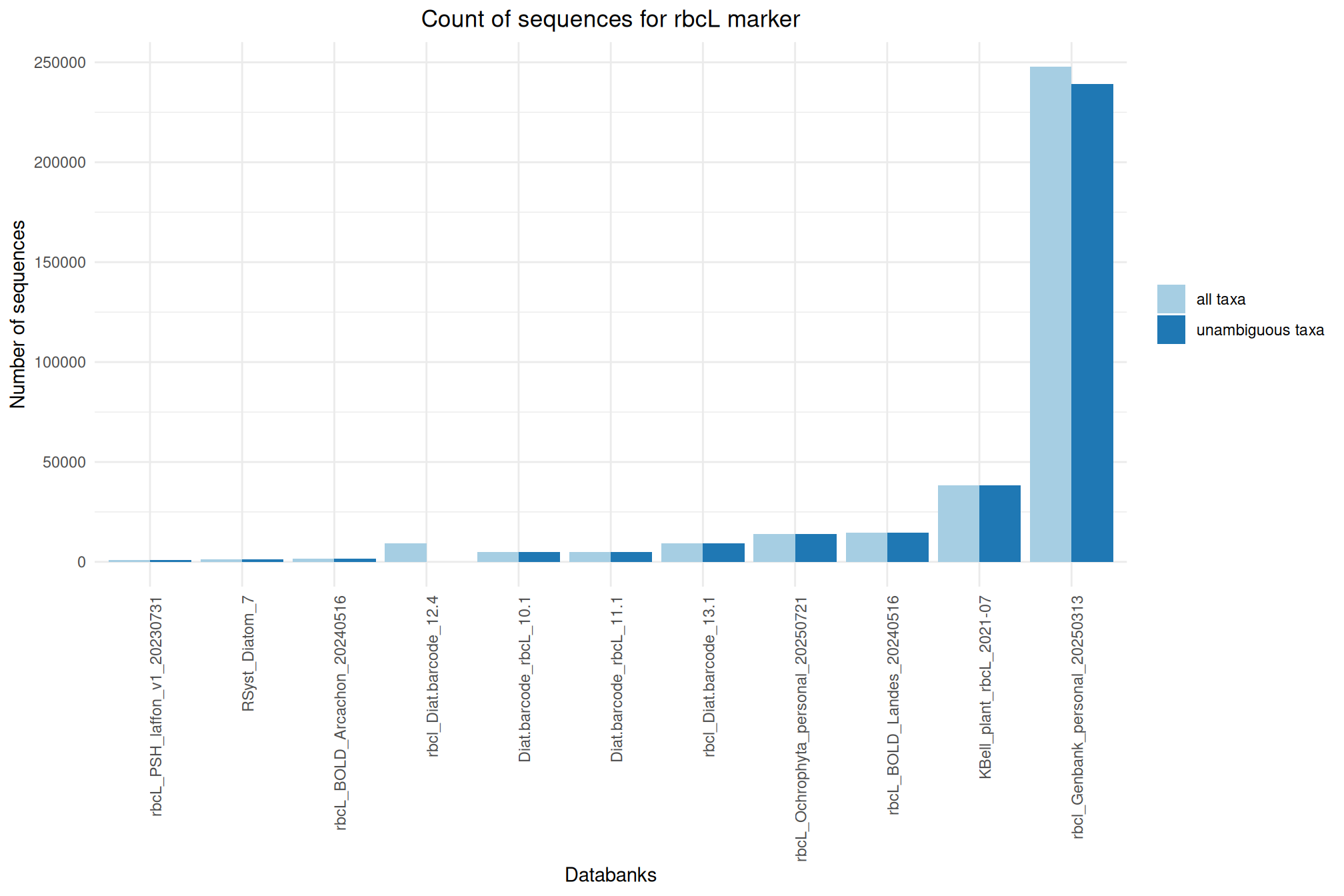
| rbcL_Ochrophyta_personal_20250721 | Jul 1, 2025 | rbcL | 13836 |
| rbcl_Diat.barcode_13.1 | Jun 1, 2025 | rbcL | 9273 |
| ITS_UNITE_Eukayote_10.0_20250219 | Apr 1, 2025 | ITS | 266589 |
| ITS_UNITE_Fungi_10.0_20250219 | Apr 1, 2025 | ITS | 168030 |
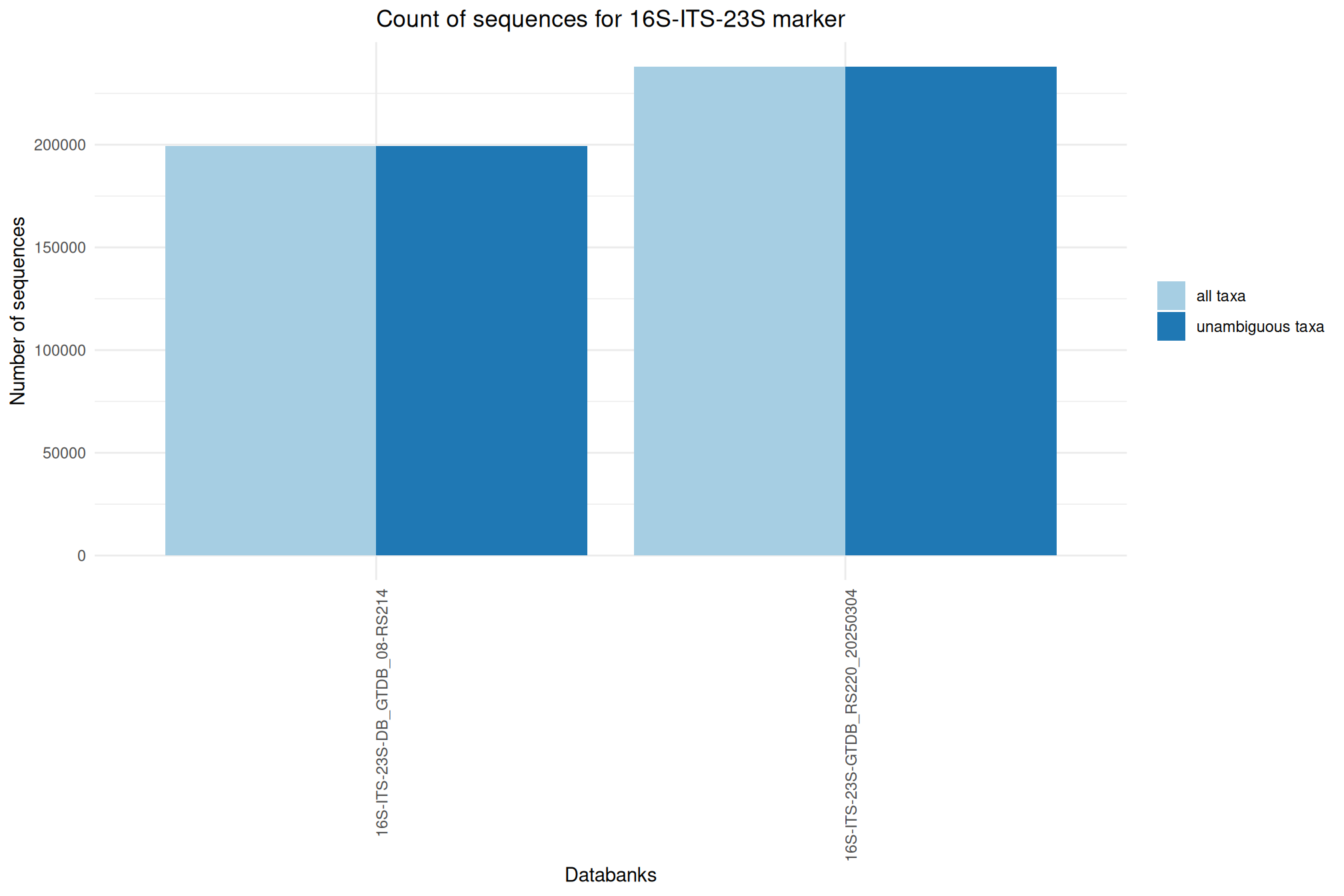
| 16S-ITS-23S-GTDB_RS220_20250304 | Mar 1, 2025 | 16S-ITS-23S | 238028 |
| rbcl_Genbank_personal_20250313 | Mar 1, 2025 | rbcL | 247718 |
| NCBIdb_archaea_16S_v2_20250204 | Feb 1, 2025 | 16S | 1149 |
| NCBIdb_bacteria_16S_v2_20250204 | Feb 1, 2025 | 16S | 26170 |
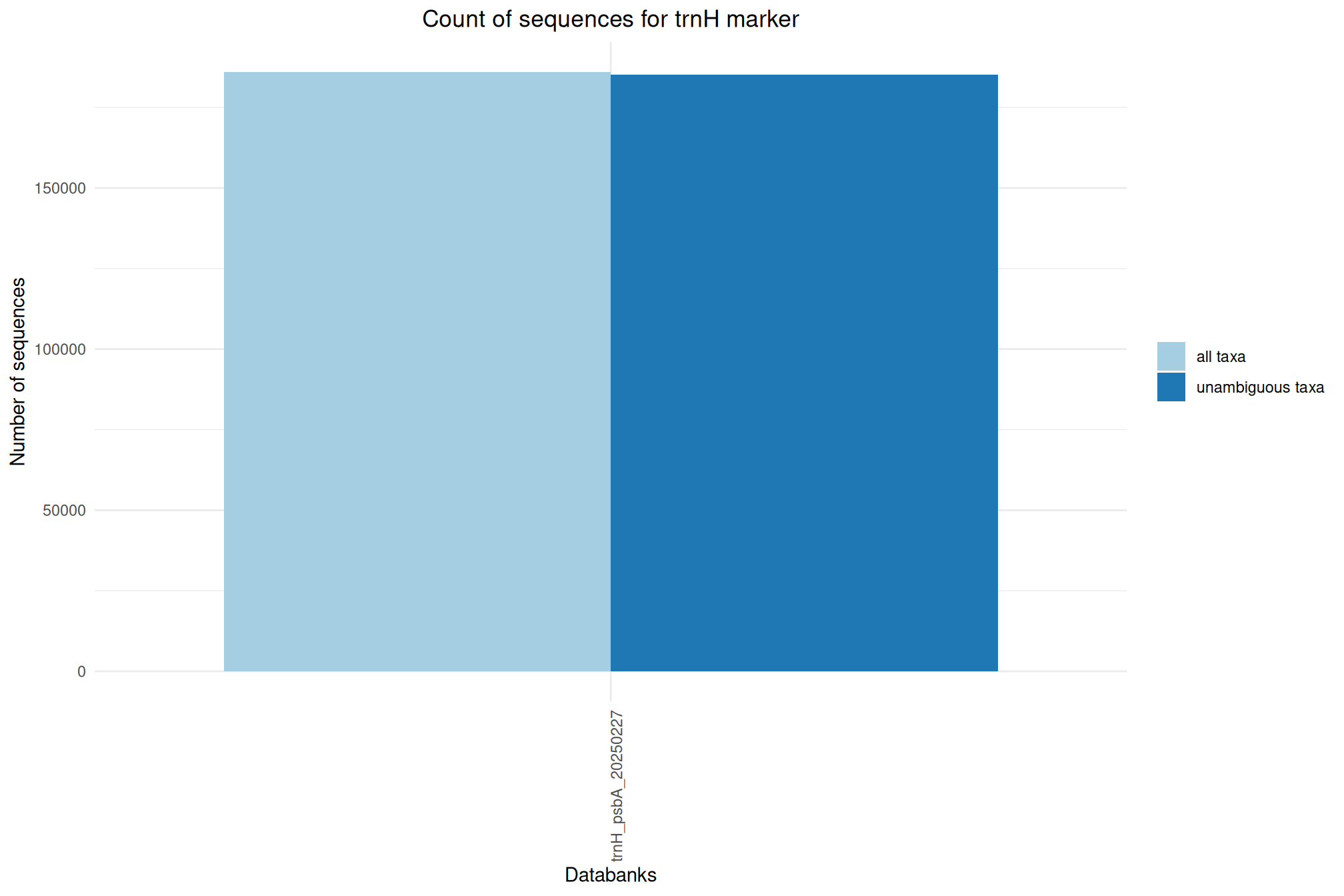
| trnH_psbA_20250227 | Feb 1, 2025 | trnH | 185964 |
| 16S_GTDB_220 | Nov 1, 2024 | 16S | 863832 |
| CyanoSeq_V1.3 | Oct 1, 2024 | 16S | 4291 |
| rbcL_Diat.barcode_12.4 | Oct 1, 2024 | rbcL | 9226 |
| 16S_SILVA_138.2 | Oct 1, 2024 | 16S | 451555 |
| 16S_SILVA_Pintail100_138.2 | Oct 1, 2024 | 16S | 317241 |
| 16S_SILVA_Pintail50_138.2 | Oct 1, 2024 | 16S | 385166 |
| 16S_SILVA_Pintail80_138.2 | Oct 1, 2024 | 16S | 366036 |
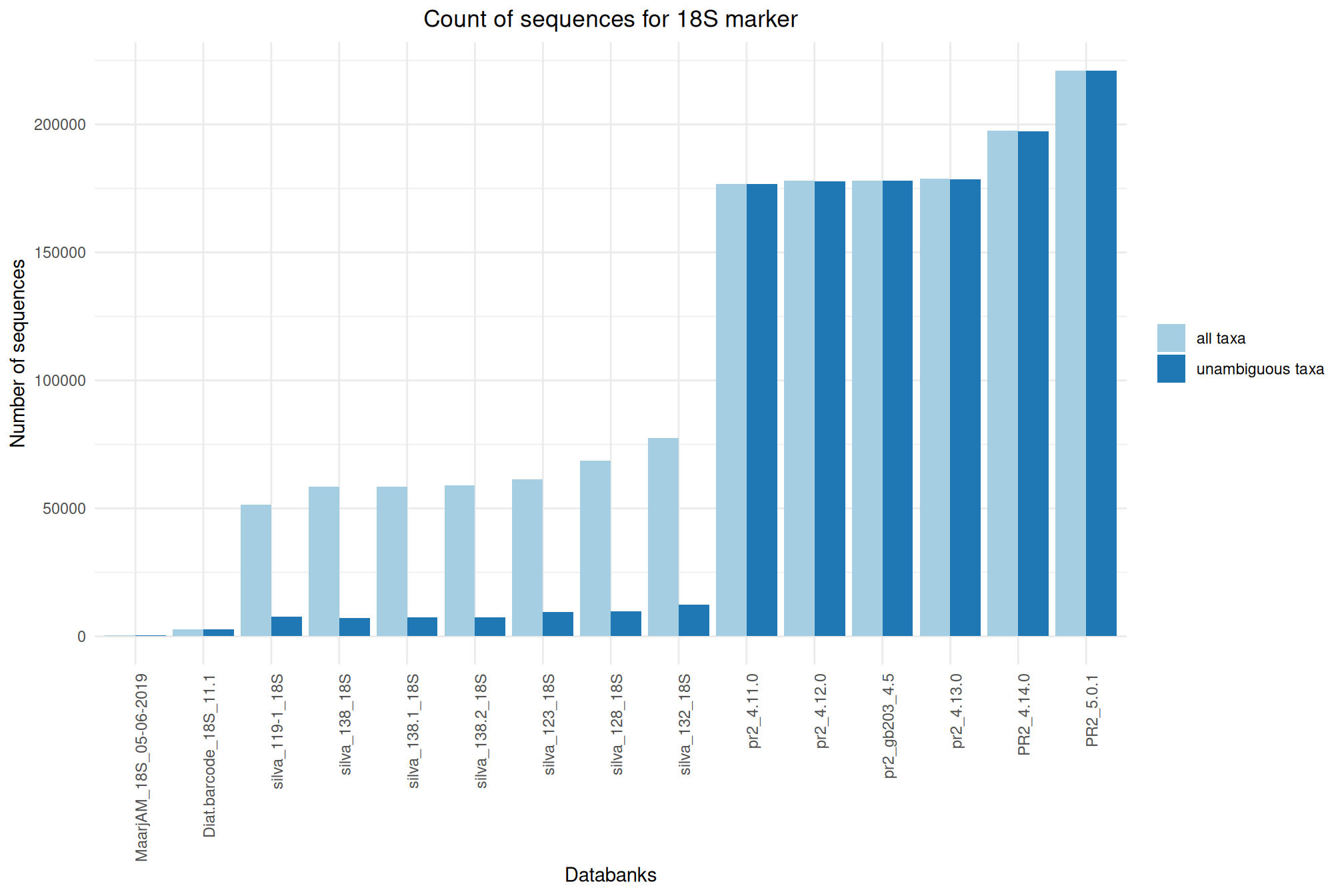
| 18S_SILVA_138.2 | Oct 1, 2024 | 18S | 58940 |
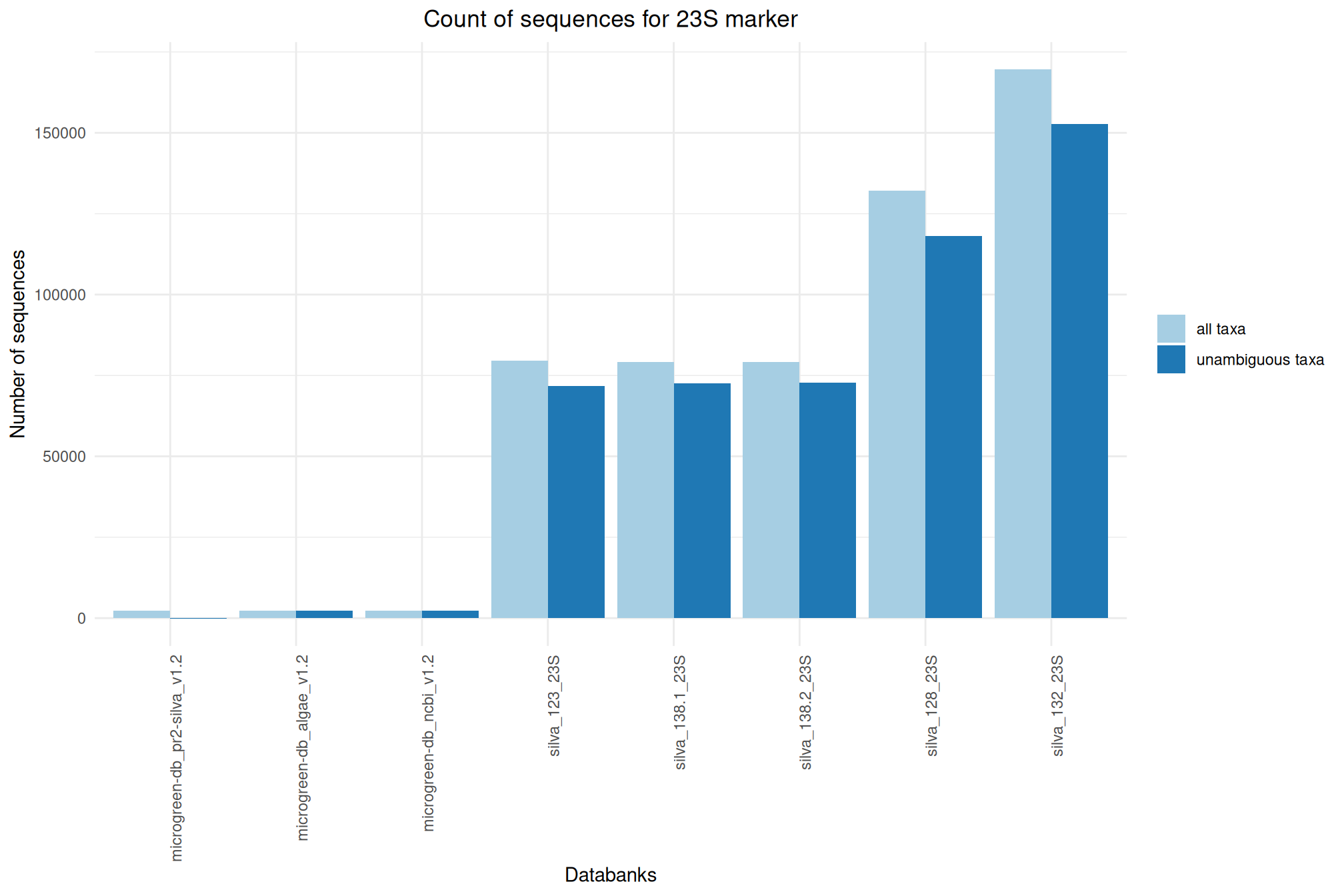
| 23S_SILVA_138.2 | Oct 1, 2024 | 23S | 79152 |
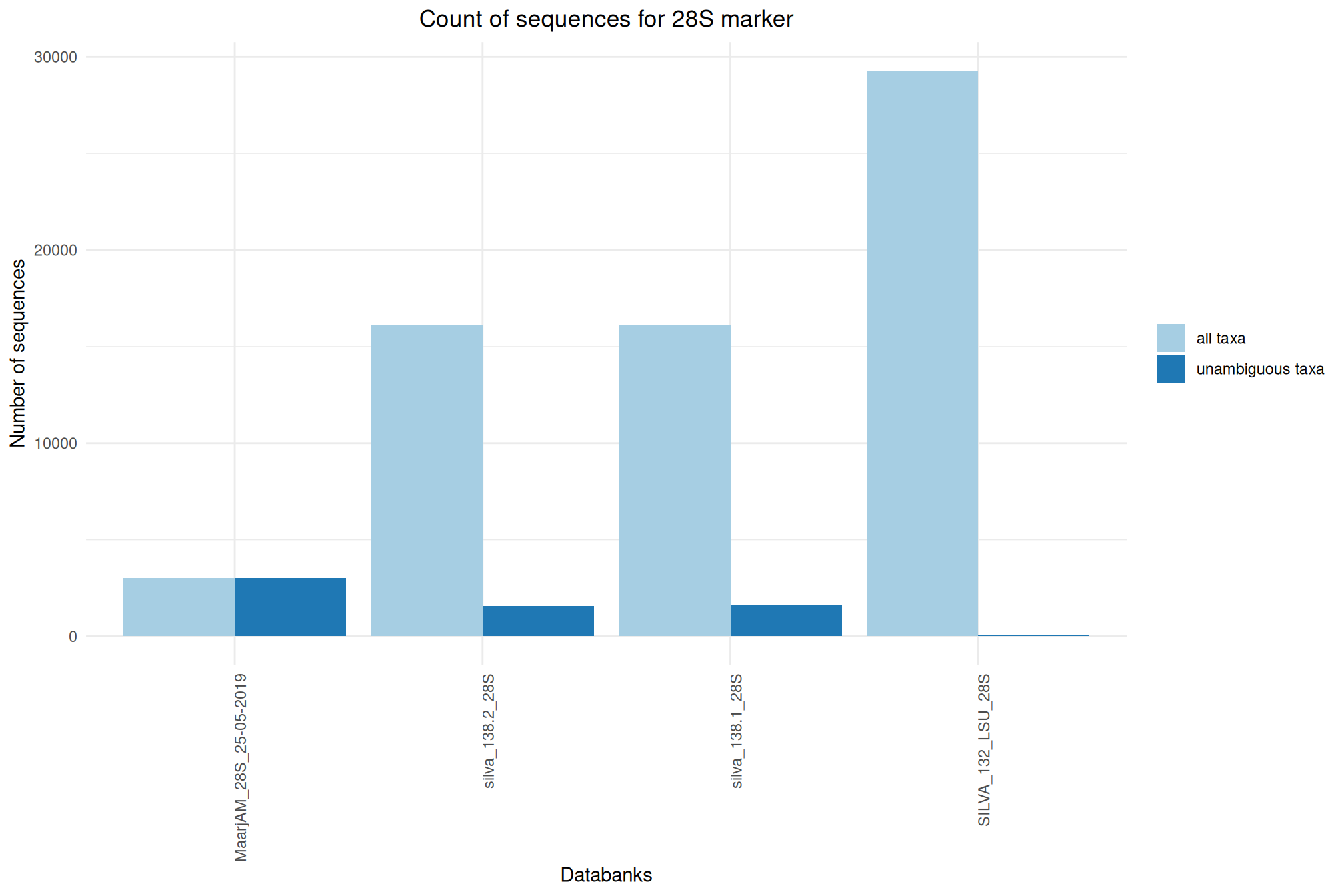
| 28S_SILVA_138.2 | Oct 1, 2024 | 28S | 16127 |
| ITS_UNITE_Eukayote_10.0 | Sep 1, 2024 | ITS | 252239 |
| ITS_UNITE_Fungi_10.0 | Sep 1, 2024 | ITS | 159195 |
| ITS_UNITE_Fungi_known_genus_10.0 | Sep 1, 2024 | ITS | 96376 |
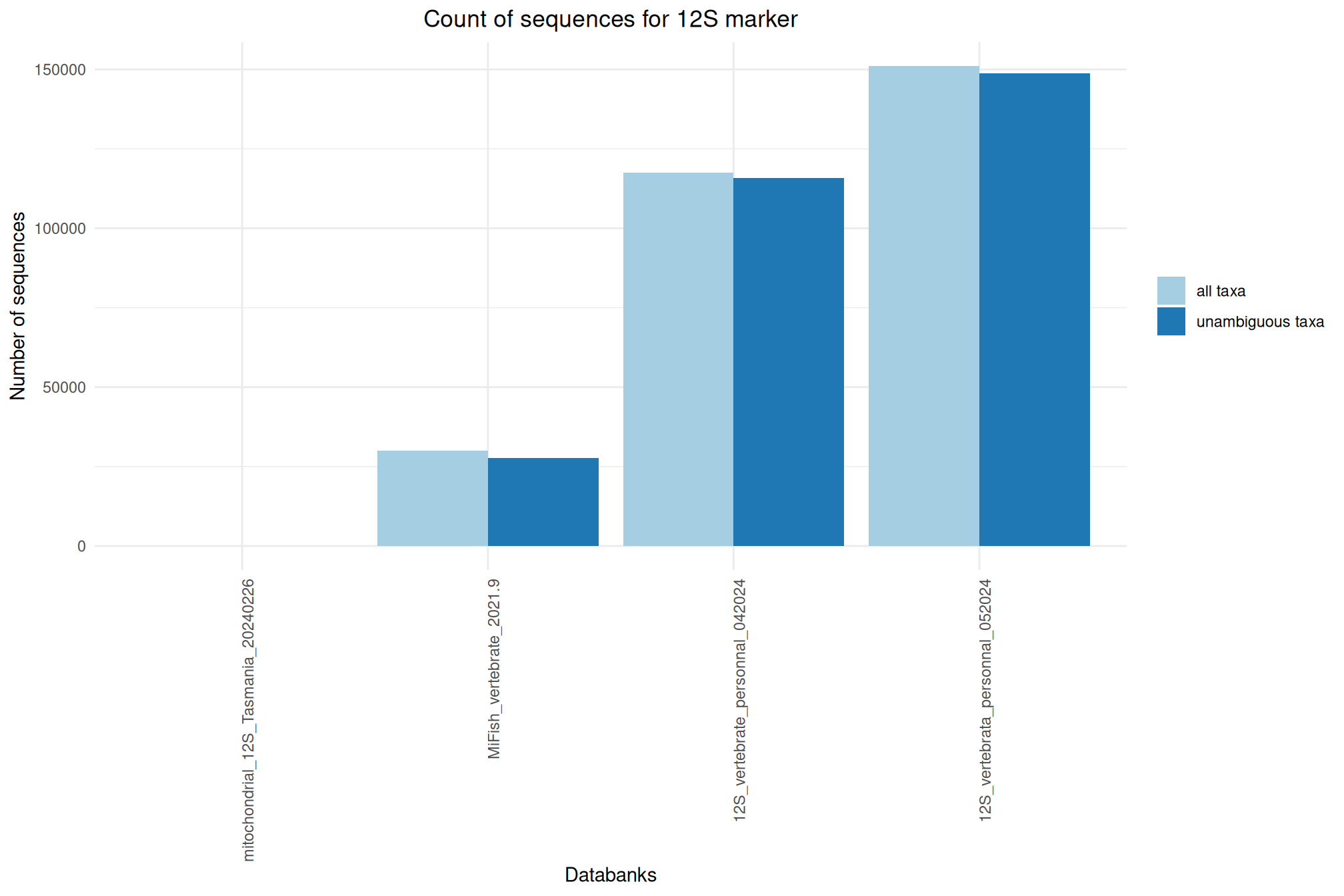
| 12S_vertebrata_personnal_052024 | Jul 1, 2024 | 12S | 151043 |
| 16S_personnal_WILDBEES_OCCITANIA_v2024071 | Jul 1, 2024 | 16S | 349 |
| COI_arthropoda_personnal_052024 | Jul 1, 2024 | COI | 52904 |
| MiFish_vertebrate_2021.9 | Jul 1, 2024 | 12S | 29938 |
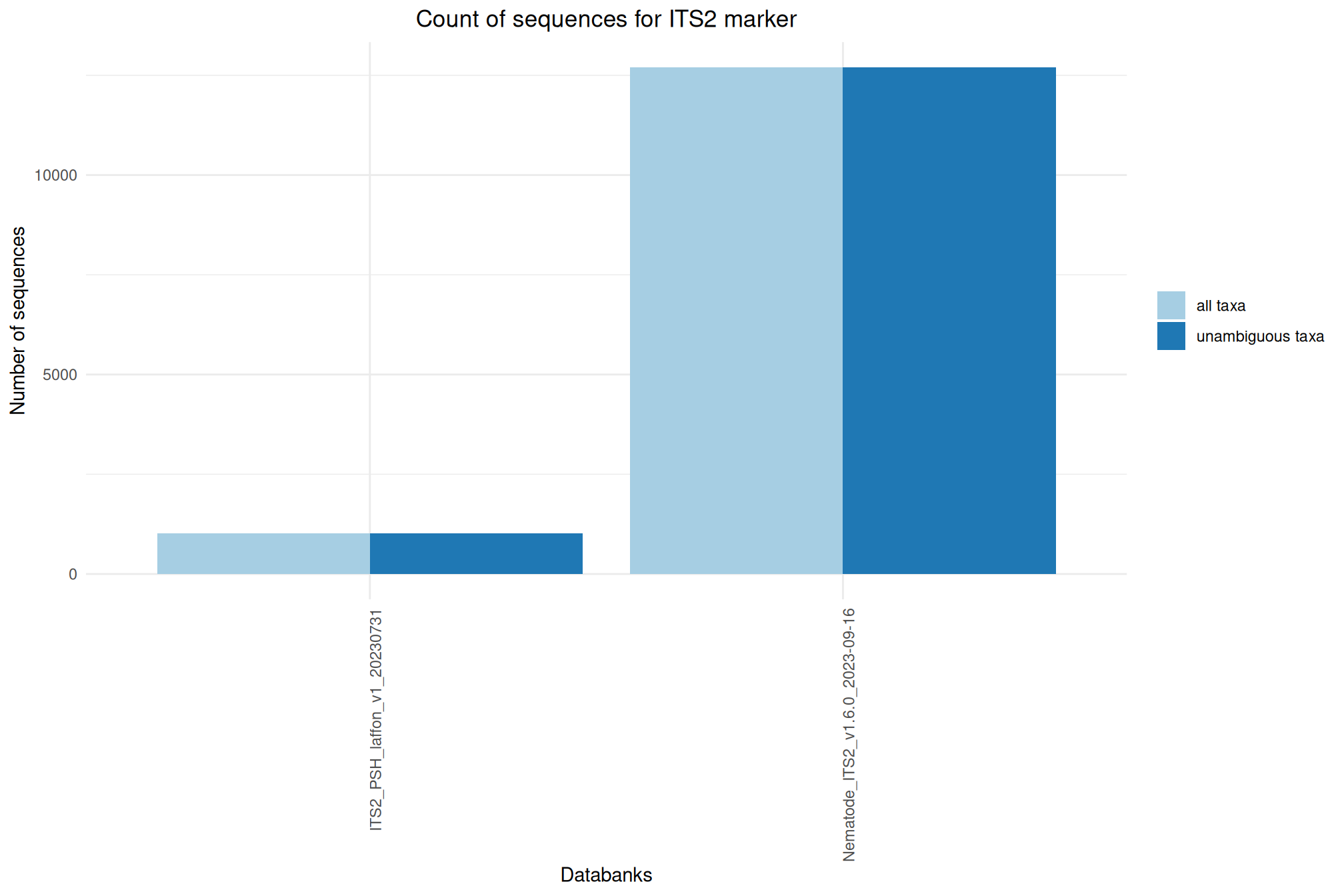
| ITS2_Nemabiome_1.6 | Jul 1, 2024 | ITS2 | 12697 |
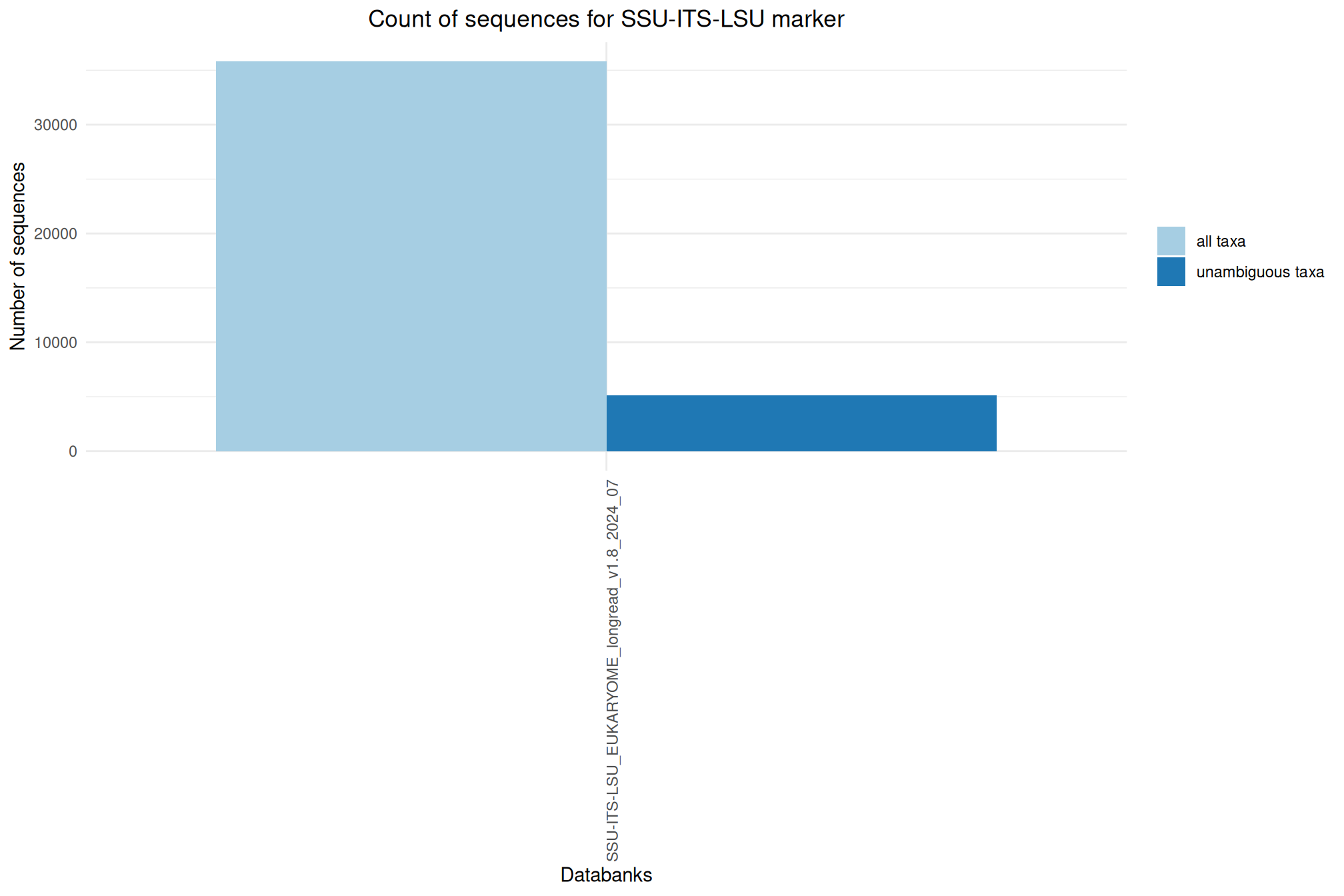
| SSU-ITS-LSU_EUKARYOME_longread_v1.8_2024_07 | Jul 1, 2024 | SSU-ITS-LSU | 35793 |
| rpoB_REFseq_bacteria_all | Jul 1, 2024 | rpoB | 362332 |
| rpoB_REFseq_bacteria_chr_and_compl_genomes | Jul 1, 2024 | rpoB | 47069 |
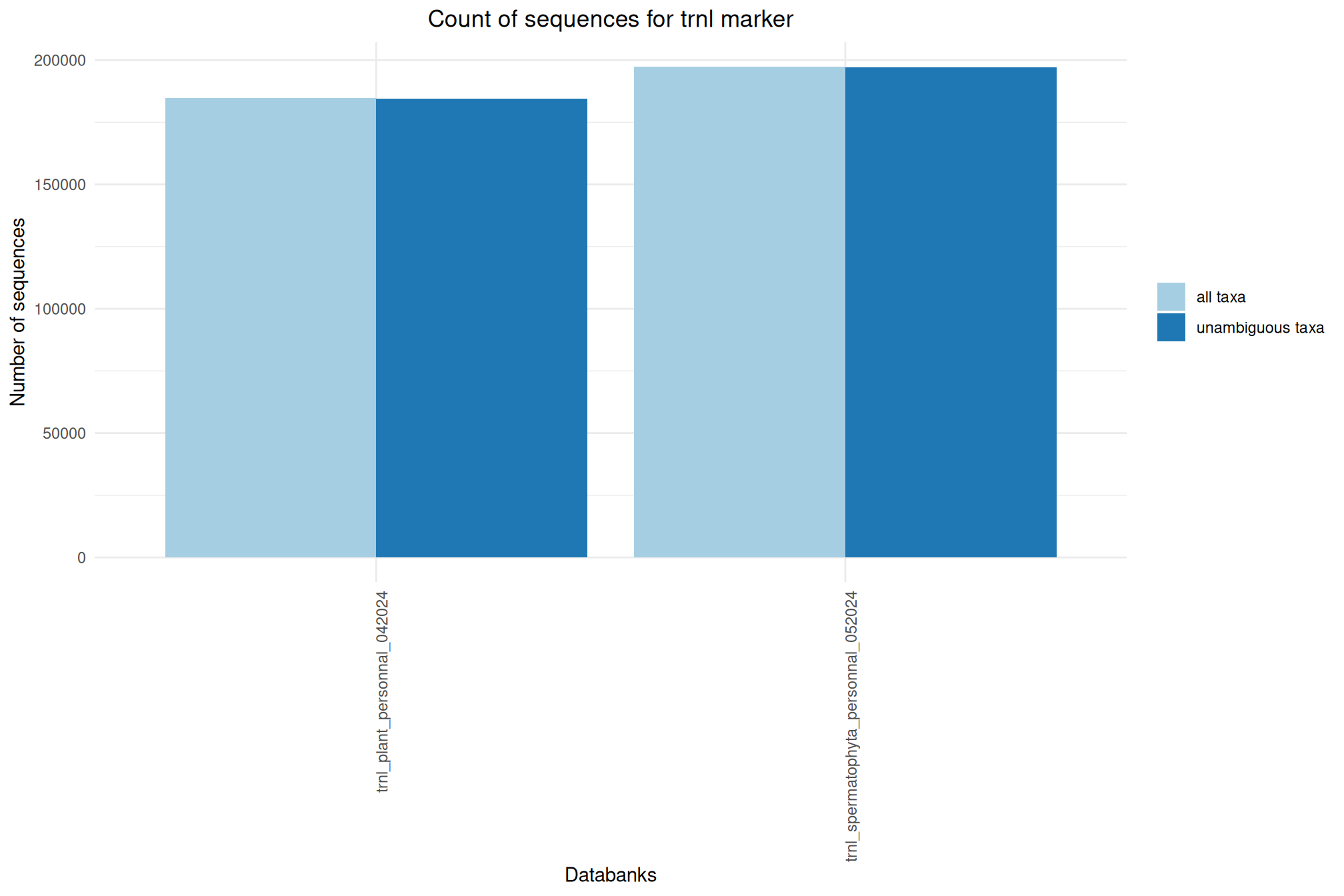
| trnl_spermatophyta_personnal_052024 | Jul 1, 2024 | trnl | 197354 |
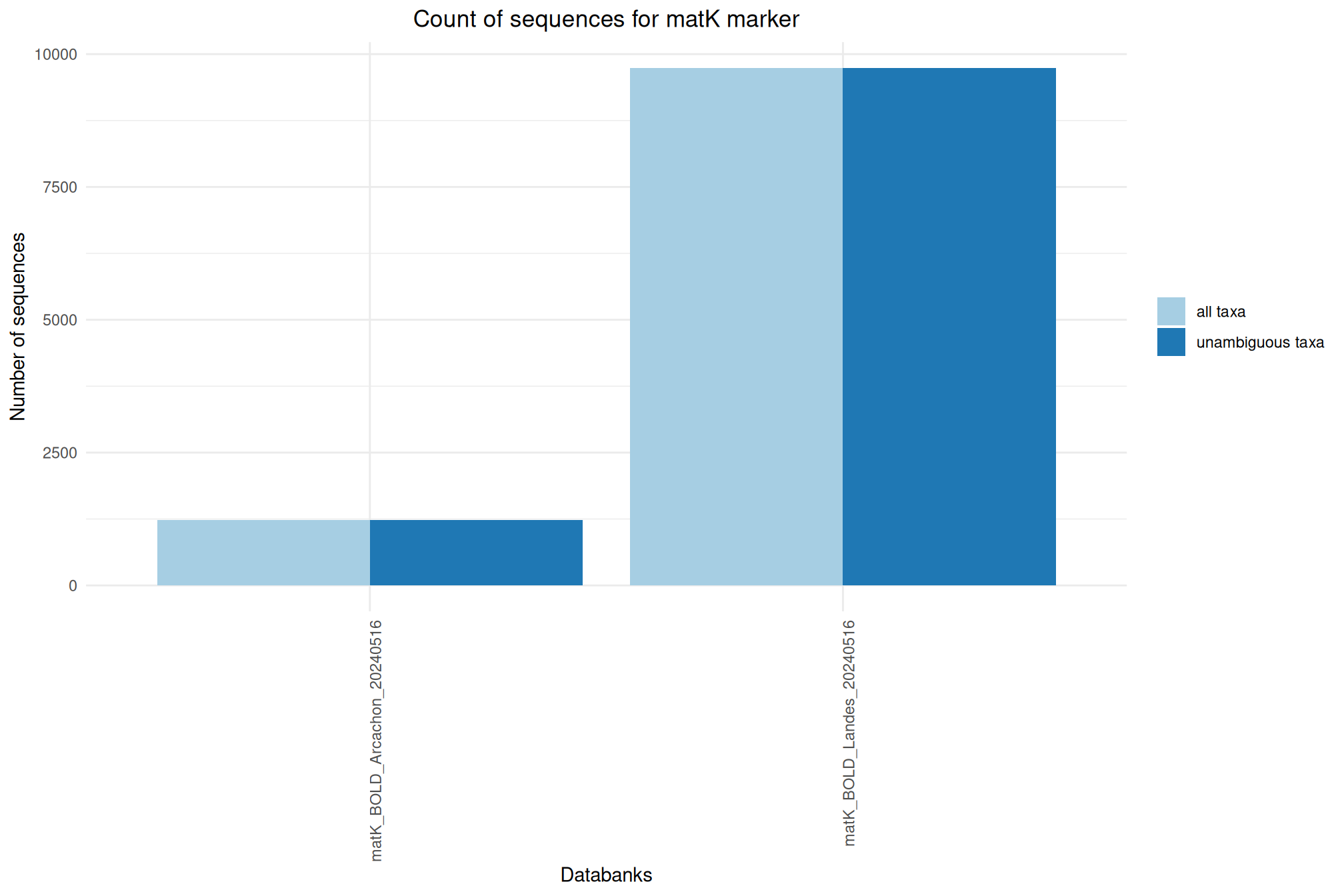
| matK_BOLD_Arcachon_20240516 | May 1, 2024 | matK | 1225 |
| matK_BOLD_Landes_20240516 | May 1, 2024 | matK | 9738 |
| rbcL_BOLD_Arcachon_20240516 | May 1, 2024 | rbcL | 1508 |
| rbcL_BOLD_Landes_20240516 | May 1, 2024 | rbcL | 14465 |
| 12S_vertebrate_personnal_042024 | Apr 1, 2024 | 12S | 117581 |
| trnl_plant_personnal_042024 | Apr 1, 2024 | trnl | 184654 |
| mitochondrial_12S_Tasmania_20240226 | Feb 1, 2024 | 12S | 52 |
| 16S_MIDAS_5.1 | Dec 1, 2023 | 16S | 120408 |
| 16S_MIDAS_5.0 | Nov 1, 2023 | 16S | 120408 |
| 16S-ITS-23S_GTDB_08-RS214 | Sep 1, 2023 | 16S-ITS-23S | 199407 |
| COI_BOLD_marin_20230913 | Sep 1, 2023 | COI | 1143010 |
| 18S_PR2_5.0.1 | Sep 1, 2023 | 18S | 221085 |
| COI_BOLD_082023 | Aug 1, 2023 | COI | 4808391 |
| ITS2_PSH_laffon_v1_20230731 | Jul 1, 2023 | ITS2 | 1013 |
| 16S_REFseq_Bacteria_20230726 | Jul 1, 2023 | 16S | 25849 |
| rbcL_PSH_laffon_v1_20230731 | Jul 1, 2023 | rbcL | 1093 |
| 16S_REFseq_Archaea_20230726 | Mar 1, 2023 | 16S | 1130 |
| ITS_UNITE_Eukayote_9.0 | Mar 1, 2023 | ITS | 326434 |
| ITS_UNITE_Fungi_9.0 | Mar 1, 2023 | ITS | 206184 |
| COI_COInr_2022_05_06 | Feb 1, 2023 | COI | 3259654 |
| 18S_Diat.barcode_11.1 | Feb 1, 2023 | 18S | 2646 |
| rbcL_Diat.barcode_11.1 | Feb 1, 2023 | rbcL | 4823 |
| COI_MIDORI2_LONGEST_SP_GB253 | Feb 1, 2023 | COI | 210210 |
| COI_MIDORI2_UNIQ_SP_GB253 | Feb 1, 2023 | COI | 1537155 |
| 23S_microgreen-db_algae_v1.2 | Feb 1, 2023 | 23S | 2326 |
| 23S_microgreen-db_ncbi_v1.2 | Feb 1, 2023 | 23S | 2326 |
| 23S_microgreen-db_pr2-silva_v1.2 | Feb 1, 2023 | 23S | 2217 |
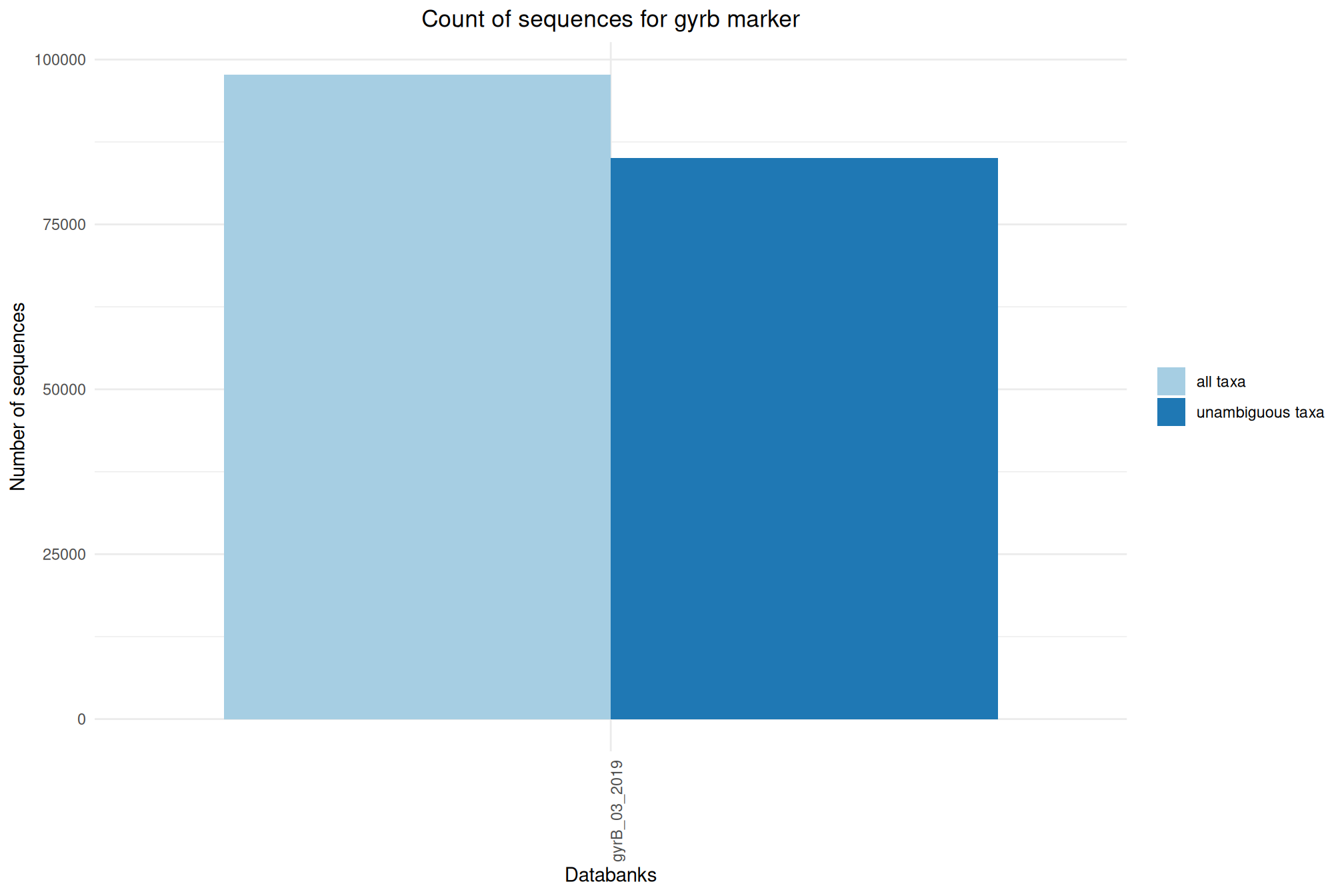
| gyrb_GyrB_03-2019 | Sep 1, 2022 | gyrb | 97747 |
| 28S_MaarjAM_25-05-2019 | Aug 1, 2022 | 28S | 3026 |
| COI_BOLD_marin_052022 | May 1, 2022 | COI | 1212507 |
| 16S_MIDAS_S138.1_v4.8.1 | May 1, 2022 | 16S | 90164 |
| COI_MIDORI_LONGEST_SP_GB249 | May 1, 2022 | COI | 785468 |
| COI_MIDORI_UNIQ_SP_GB249 | May 1, 2022 | COI | 2651489 |
| 18S_PR2_4.14.0 | May 1, 2022 | 18S | 197602 |
| rbcL_KBell_plant_2021-07 | Jan 1, 2022 | rbcL | 38464 |
| 16S_DAIRYdb_v1.2.4_20200604 | Dec 1, 2021 | 16S | 10439 |
| 16S_DAIRYdb_v2.0_20210401 | Dec 1, 2021 | 16S | 10611 |
| ITS_UNITE_Eukaryote_8.0 | Dec 1, 2021 | ITS | 205125 |
| ITS_UNITE_Fungi_8.3 | Dec 1, 2021 | ITS | 98090 |
| rbcL_Diat.barcode_10.1 | Nov 1, 2021 | rbcL | 4798 |
| COI_MIDORI_LONGEST_SP_GB242 | Jun 1, 2021 | COI | 746139 |
| 18S_PR2_4.13.0 | May 1, 2021 | 18S | 178794 |
| 18S_MaarjAM_05-06-201 | Mar 1, 2021 | 18S | 385 |
| 16S_SILVA_138.1 | Sep 1, 2020 | 16S | 451964 |
| 16S_SILVA_Pintail100_138.1 | Sep 1, 2020 | 16S | 317435 |
| 16S_SILVA_Pintail50_138.1 | Sep 1, 2020 | 16S | 385381 |
| 16S_SILVA_Pintail80_138.1 | Sep 1, 2020 | 16S | 366234 |
| 18S_SILVA_138.1 | Sep 1, 2020 | 18S | 58544 |
| 23S_SILVA_138.1 | Sep 1, 2020 | 23S | 79135 |
| 28S_SILVA_138.1 | Sep 1, 2020 | 28S | 16151 |
| 16S_MIDAS_S132_3.6 | Mar 1, 2020 | 16S | 9609 |
| ITS_UNITE_Eukaryote_8.2 | Mar 1, 2020 | ITS | 196344 |
| ITS_UNITE_Fungi_8.2 | Mar 1, 2020 | ITS | 84387 |
| 28S_SILVA_132 | Feb 1, 2020 | 28S | 29298 |
| 16S_SILVA_138 | Feb 1, 2020 | 16S | 452422 |
| 16S_SILVA_Pintail100_138 | Feb 1, 2020 | 16S | 317680 |
| 16S_SILVA_Pintail50_138 | Feb 1, 2020 | 16S | 385631 |
| 16S_SILVA_Pintail80_138 | Feb 1, 2020 | 16S | 366234 |
| 18S_SILVA_138 | Feb 1, 2020 | 18S | 58562 |
| 18S_PR2_4.12.0 | Jan 1, 2020 | 18S | 177934 |
| COI_MIDORI_MARINE_20180221 | Aug 1, 2019 | COI | 335177 |
| COI_MIDORI_20180221 | Jul 1, 2019 | COI | 927386 |
| ITS_UNITE_Eukaryote_8.0 | Mar 1, 2019 | ITS | 122187 |
| ITS_UNITE_Fungi_8.0 | Mar 1, 2019 | ITS | 71042 |
| COI_BOLD_22019 | Feb 1, 2019 | COI | 2276871 |
| COI_BOLD_1percentN_22019 | Feb 1, 2019 | COI | 2206551 |
| 16S_DAIRYdb_V1.1.2 | Dec 1, 2018 | 16S | 10290 |
| 16S_EZBioCloud_52018 | Dec 1, 2018 | 16S | 64660 |
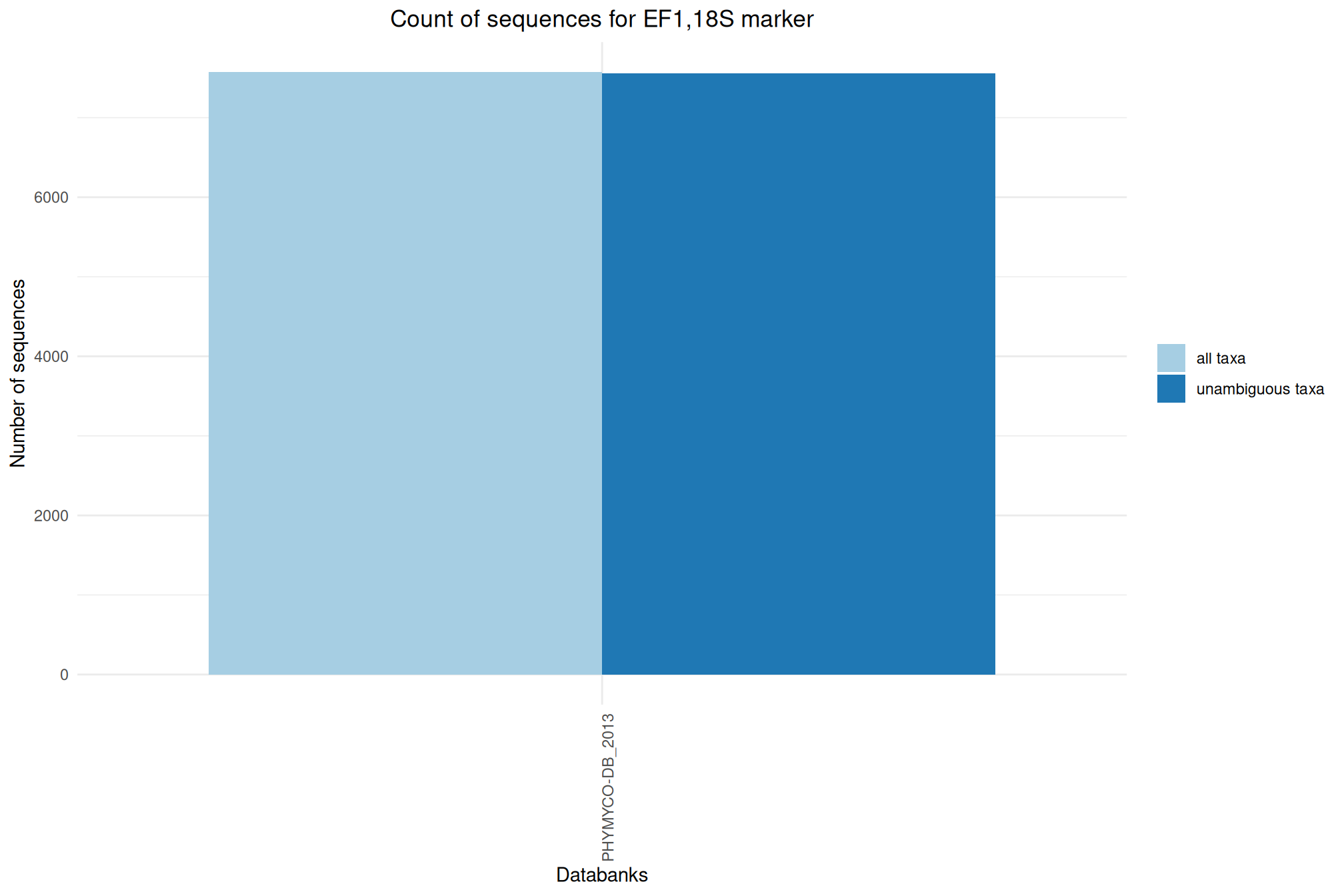
| EF1-18S_PHYMYCO-DB_2013 | Dec 1, 2018 | EF1,18S | 7571 |
| rbcL_Rsyst_Diatom_7 | Dec 1, 2018 | rbcL | 1401 |
| 18S_PR2_4.11.0 | Dec 1, 2018 | 18S | 176818 |
| ITS_UNITE_7.1 | Oct 1, 2018 | ITS | 54568 |
| rpoB_INRA_122017 | Jul 1, 2018 | rpoB | 44673 |
| 16S_SILVA_132 | Feb 1, 2018 | 16S | 617631 |
| 16S_SILVA_Pintail100_132 | Feb 1, 2018 | 16S | 476070 |
| 16S_SILVA_Pintail50_132 | Feb 1, 2018 | 16S | 580207 |
| 16S_SILVA_Pintail80_132 | Feb 1, 2018 | 16S | 558585 |
| 18S_SILVA_132 | Feb 1, 2018 | 18S | 77540 |
| 23S_SILVA_132 | Feb 1, 2018 | 23S | 169545 |
| 16S_SILVA_Pintail100_128 | Jul 1, 2017 | 16S | 440703 |
| 16S_SILVA_Pintail50_128 | Jul 1, 2017 | 16S | 521508 |
| 16S_SILVA_Pintail80_128 | Jul 1, 2017 | 16S | 500466 |
| 16S_MIDAS_S123_2.1.3 | Jan 1, 2017 | 16S | 534966 |
| 18S_PR2_Gb203_4.5 | Jan 1, 2017 | 18S | 178094 |
| 18S_SILVA_123 | Dec 1, 2016 | 18S | 61384 |
| 16S_SILVA_128 | Dec 1, 2016 | 16S | 576542 |
| 18S_SILVA_128 | Dec 1, 2016 | 18S | 68609 |
| 23S_SILVA_128 | Dec 1, 2016 | 23S | 132194 |
| 16S_MIDAS_S119_1.20 | Feb 1, 2016 | 16S | 463278 |
| 16S_Greengenes_13.5 | Feb 1, 2016 | 16S | 1262986 |
| 18S_SILVA_119-1 | Jan 1, 2016 | 18S | 51553 |
| 16S_SILVA_123 | Jan 1, 2016 | 16S | 536223 |
| 23S_SILVA_123 | Jan 1, 2016 | 23S | 79661 |
No matching items
Ratio of unambiguous sequences
A significant number of sequences in databanks have a taxonomy non informative. Ambiguous taxa contains; metagenome, unknown, unspecified, unidentified, unclassified, incertae, NA, None.